إنزيم اقتطاع
The cutting of DNA at specific boundaries
Used by restriction enzymes to locate specific sequences of DNA on which to bind and subsequently cleave
The DNA sequence to which restriction enzymes bind
The DNA sequence that is cleaved by the restriction enzyme
إنزيمات الاقتطاع إنگليزية: restiction enzymes restriction endonuclease, REase, ENase or restrictase هي الإنزيمات التي تقطع تسلسلات الدنا عند مواضع محددة تعرف بمواقع الاقتطاع restriction site.[1][2][3] Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each sugar-phosphate backbone (i.e. each strand) of the DNA double helix.
وتوجد إنزيمات الاقتطاع لدى الجراثيم الذي يعتقد بأنه يقيها من غزو العاثيات وتستخدم تلك الإنزيمات كأدوات في الدراسات الوراثية والتشخيص الوراثي.
These enzymes are found in bacteria and archaea and provide a defense mechanism against invading viruses.[4][5] Inside a prokaryote, the restriction enzymes selectively cut up foreign DNA in a process called restriction digestion; meanwhile, host DNA is protected by a modification enzyme (a methyltransferase) that modifies the prokaryotic DNA and blocks cleavage. Together, these two processes form the restriction modification system.[6]
More than 3,600 restriction endonucleases are known which represent over 250 different specificities.[7] Over 3,000 of these have been studied in detail, and more than 800 of these are available commercially.[8] These enzymes are routinely used for DNA modification in laboratories, and they are a vital tool in molecular cloning.[9][10][11]
التاريخ
The term restriction enzyme originated from the studies of phage λ, a virus that infects bacteria, and the phenomenon of host-controlled restriction and modification of such bacterial phage or bacteriophage.[12] The phenomenon was first identified in work done in the laboratories of Salvador Luria, Jean Weigle and Giuseppe Bertani in the early 1950s.[13][14] It was found that, for a bacteriophage λ that can grow well in one strain of Escherichia coli, for example E. coli C, when grown in another strain, for example E. coli K, its yields can drop significantly, by as much as three to five orders of magnitude. The host cell, in this example E. coli K, is known as the restricting host and appears to have the ability to reduce the biological activity of the phage λ. If a phage becomes established in one strain, the ability of that phage to grow also becomes restricted in other strains. In the 1960s, it was shown in work done in the laboratories of Werner Arber and Matthew Meselson that the restriction is caused by an enzymatic cleavage of the phage DNA, and the enzyme involved was therefore termed a restriction enzyme.[4][15][16][17]
The restriction enzymes studied by Arber and Meselson were type I restriction enzymes, which cleave DNA randomly away from the recognition site.[18] In 1970, Hamilton O. Smith, Thomas Kelly and Kent Wilcox isolated and characterized the first type II restriction enzyme, HindII, from the bacterium Haemophilus influenzae.[19][20] Restriction enzymes of this type are more useful for laboratory work as they cleave DNA at the site of their recognition sequence and are the most commonly used as a molecular biology tool.[21] Later, Daniel Nathans and Kathleen Danna showed that cleavage of simian virus 40 (SV40) DNA by restriction enzymes yields specific fragments that can be separated using polyacrylamide gel electrophoresis, thus showing that restriction enzymes can also be used for mapping DNA.[22] For their work in the discovery and characterization of restriction enzymes, the 1978 Nobel Prize for Physiology or Medicine was awarded to Werner Arber, Daniel Nathans, and Hamilton O. Smith.[23] The discovery of restriction enzymes allows DNA to be manipulated, leading to the development of recombinant DNA technology that has many applications, for example, allowing the large scale production of proteins such as human insulin used by diabetic patients.[13][24]
الأصول
Restriction enzymes likely evolved from a common ancestor and became widespread via horizontal gene transfer.[25][26] In addition, there is mounting evidence that restriction endonucleases evolved as a selfish genetic element.[27]
موقع التعرف
Restriction enzymes recognize a specific sequence of nucleotides[2] and produce a double-stranded cut in the DNA. The recognition sequences can also be classified by the number of bases in its recognition site, usually between 4 and 8 bases, and the number of bases in the sequence will determine how often the site will appear by chance in any given genome, e.g., a 4-base pair sequence would theoretically occur once every 4^4 or 256bp, 6 bases, 4^6 or 4,096bp, and 8 bases would be 4^8 or 65,536bp.[28] Many of them are palindromic, meaning the base sequence reads the same backwards and forwards.[29] In theory, there are two types of palindromic sequences that can be possible in DNA. The mirror-like palindrome is similar to those found in ordinary text, in which a sequence reads the same forward and backward on a single strand of DNA, as in GTAATG. The inverted repeat palindrome is also a sequence that reads the same forward and backward, but the forward and backward sequences are found in complementary DNA strands (i.e., of double-stranded DNA), as in GTATAC (GTATAC being complementary to CATATG).[30] Inverted repeat palindromes are more common and have greater biological importance than mirror-like palindromes.
EcoRI digestion produces "sticky" ends,
whereas SmaI restriction enzyme cleavage produces "blunt" ends:
Recognition sequences in DNA differ for each restriction enzyme, producing differences in the length, sequence and strand orientation (5' end or 3' end) of a sticky-end "overhang" of an enzyme restriction.[31]
Different restriction enzymes that recognize the same sequence are known as neoschizomers. These often cleave in different locales of the sequence. Different enzymes that recognize and cleave in the same location are known as isoschizomers.
الأنواع
Naturally occurring restriction endonucleases are categorized into five groups (Types I, II, III, IV, and V) based on their composition and enzyme cofactor requirements, the nature of their target sequence, and the position of their DNA cleavage site relative to the target sequence.[32][33][34] DNA sequence analysis of restriction enzymes however show great variations, indicating that there are more than four types.[35] All types of enzymes recognize specific short DNA sequences and carry out the endonucleolytic cleavage of DNA to give specific fragments with terminal 5'-phosphates. They differ in their recognition sequence, subunit composition, cleavage position, and cofactor requirements,[36][37] as summarised below:
- Type I enzymes (
EC 3.1.21.3 ) cleave at sites remote from a recognition site; require both ATP and S-adenosyl-L-methionine to function; multifunctional protein with both restriction digestion and methylase ( EC 2.1.1.72 ) activities.
- Type II enzymes (
EC 3.1.21.4 ) cleave within or at short specific distances from a recognition site; most require magnesium; single function (restriction digestion) enzymes independent of methylase.
- Type III enzymes (
EC 3.1.21.5 ) cleave at sites a short distance from a recognition site; require ATP (but do not hydrolyse it); S-adenosyl-L-methionine stimulates the reaction but is not required; exist as part of a complex with a modification methylase ( EC 2.1.1.72 ).
- Type IV enzymes target modified DNA, e.g. methylated, hydroxymethylated and glucosyl-hydroxymethylated DNA
- Type V enzymes utilize guide RNAs (gRNAs)
النوع I
Type I restriction enzymes were the first to be identified and were first identified in two different strains (K-12 and B) of E. coli.[38] These enzymes cut at a site that differs, and is a random distance (at least 1000 bp) away, from their recognition site. Cleavage at these random sites follows a process of DNA translocation, which shows that these enzymes are also molecular motors. The recognition site is asymmetrical and is composed of two specific portions—one containing 3–4 nucleotides, and another containing 4–5 nucleotides—separated by a non-specific spacer of about 6–8 nucleotides. These enzymes are multifunctional and are capable of both restriction digestion and modification activities, depending upon the methylation status of the target DNA. The cofactors S-Adenosyl methionine (AdoMet), hydrolyzed adenosine triphosphate (ATP), and magnesium (Mg2+) ions, are required for their full activity. Type I restriction enzymes possess three subunits called HsdR, HsdM, and HsdS; HsdR is required for restriction digestion; HsdM is necessary for adding methyl groups to host DNA (methyltransferase activity), and HsdS is important for specificity of the recognition (DNA-binding) site in addition to both restriction digestion (DNA cleavage) and modification (DNA methyltransferase) activity.[32][38]
النوع II
| Type II site-specific deoxyribonuclease-like | |
|---|---|
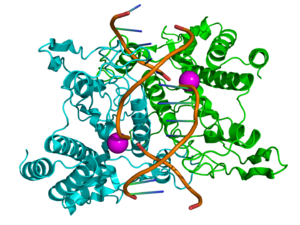 Structure of the homodimeric restriction enzyme EcoRI (cyan and green cartoon diagram) bound to double stranded DNA (brown tubes).[39] Two catalytic magnesium ions (one from each monomer) are shown as magenta spheres and are adjacent to the cleaved sites in the DNA made by the enzyme (depicted as gaps in the DNA backbone). | |
| المعرفات | |
| الرمز | Restrct_endonuc-II-like |
| عشيرة Pfam | CL0236 |
| InterPro | IPR011335 |
| SCOP | 1wte |
| SUPERFAMILY | 1wte |
Typical type II restriction enzymes differ from type I restriction enzymes in several ways. They form homodimers, with recognition sites that are usually undivided and palindromic and 4–8 nucleotides in length. They recognize and cleave DNA at the same site, and they do not use ATP or AdoMet for their activity—they usually require only Mg2+ as a cofactor.[29] These enzymes cleave the phosphodiester bond of double helix DNA. It can either cleave at the center of both strands to yield a blunt end, or at a staggered position leaving overhangs called sticky ends.[40] These are the most commonly available and used restriction enzymes. In the 1990s and early 2000s, new enzymes from this family were discovered that did not follow all the classical criteria of this enzyme class, and new subfamily nomenclature was developed to divide this large family into subcategories based on deviations from typical characteristics of type II enzymes.[29] These subgroups are defined using a letter suffix.
Type IIB restriction enzymes (e.g., BcgI and BplI) are multimers, containing more than one subunit.[29] They cleave DNA on both sides of their recognition to cut out the recognition site. They require both AdoMet and Mg2+ cofactors. Type IIE restriction endonucleases (e.g., NaeI) cleave DNA following interaction with two copies of their recognition sequence.[29] One recognition site acts as the target for cleavage, while the other acts as an allosteric effector that speeds up or improves the efficiency of enzyme cleavage. Similar to type IIE enzymes, type IIF restriction endonucleases (e.g. NgoMIV) interact with two copies of their recognition sequence but cleave both sequences at the same time.[29] Type IIG restriction endonucleases (e.g., RM.Eco57I) do have a single subunit, like classical Type II restriction enzymes, but require the cofactor AdoMet to be active.[29] Type IIM restriction endonucleases, such as DpnI, are able to recognize and cut methylated DNA.[29][41][42] Type IIS restriction endonucleases (e.g. FokI) cleave DNA at a defined distance from their non-palindromic asymmetric recognition sites;[29] this characteristic is widely used to perform in-vitro cloning techniques such as Golden Gate cloning. These enzymes may function as dimers. Similarly, Type IIT restriction enzymes (e.g., Bpu10I and BslI) are composed of two different subunits. Some recognize palindromic sequences while others have asymmetric recognition sites.[29]
النوع III
Type III restriction enzymes (e.g., EcoP15) recognize two separate non-palindromic sequences that are inversely oriented. They cut DNA about 20–30 base pairs after the recognition site.[43] These enzymes contain more than one subunit and require AdoMet and ATP cofactors for their roles in DNA methylation and restriction digestion, respectively.[44] They are components of prokaryotic DNA restriction-modification mechanisms that protect the organism against invading foreign DNA. Type III enzymes are hetero-oligomeric, multifunctional proteins composed of two subunits, Res (UniProt P08764) and Mod (UniProt P08763). The Mod subunit recognises the DNA sequence specific for the system and is a modification methyltransferase; as such, it is functionally equivalent to the M and S subunits of type I restriction endonuclease. Res is required for restriction digestion, although it has no enzymatic activity on its own. Type III enzymes recognise short 5–6 bp-long asymmetric DNA sequences and cleave 25–27 bp downstream to leave short, single-stranded 5' protrusions. They require the presence of two inversely oriented unmethylated recognition sites for restriction digestion to occur. These enzymes methylate only one strand of the DNA, at the N-6 position of adenine residues, so newly replicated DNA will have only one strand methylated, which is sufficient to protect against restriction digestion. Type III enzymes belong to the beta-subfamily of N6 adenine methyltransferases, containing the nine motifs that characterise this family, including motif I, the AdoMet binding pocket (FXGXG), and motif IV, the catalytic region (S/D/N (PP) Y/F).[36][45]
Type IV
Type IV enzymes recognize modified, typically methylated DNA and are exemplified by the McrBC and Mrr systems of E. coli.[35]
Type V
Type V restriction enzymes (e.g., the cas9-gRNA complex from CRISPRs[46]) utilize guide RNAs to target specific non-palindromic sequences found on invading organisms. They can cut DNA of variable length, provided that a suitable guide RNA is provided. The flexibility and ease of use of these enzymes make them promising for future genetic engineering applications.[46][47]
Artificial restriction enzymes
Artificial restriction enzymes can be generated by fusing a natural or engineered DNA-binding domain to a nuclease domain (often the cleavage domain of the type IIS restriction enzyme FokI).[48] Such artificial restriction enzymes can target large DNA sites (up to 36 bp) and can be engineered to bind to desired DNA sequences.[49] Zinc finger nucleases are the most commonly used artificial restriction enzymes and are generally used in genetic engineering applications,[50][51][52][53] but can also be used for more standard gene cloning applications.[54] Other artificial restriction enzymes are based on the DNA binding domain of TAL effectors.[55][56]
In 2013, a new technology CRISPR-Cas9, based on a prokaryotic viral defense system, was engineered for editing the genome, and it was quickly adopted in laboratories.[57] For more detail, read CRISPR (Clustered regularly interspaced short palindromic repeats).
In 2017, a group from University of Illinois reported using an Argonaute protein taken from Pyrococcus furiosus (PfAgo) along with guide DNA to edit DNA in vitro as artificial restriction enzymes.[58]
Artificial ribonucleases that act as restriction enzymes for RNA have also been developed. A PNA-based system, called a PNAzyme, has a Cu(II)-2,9-dimethylphenanthroline group that mimics ribonucleases for specific RNA sequence and cleaves at a non-base-paired region (RNA bulge) of the targeted RNA formed when the enzyme binds the RNA. This enzyme shows selectivity by cleaving only at one site that either does not have a mismatch or is kinetically preferred out of two possible cleavage sites.[59]
المصطلحات
| Derivation of the EcoRI name | ||
|---|---|---|
| Abbreviation | Meaning | Description |
| E | Escherichia | الجنس |
| co | coli | specific species |
| R | RY13 | strain |
| I | First identified | order of identification in the bacterium |
Since their discovery in the 1970s, many restriction enzymes have been identified; for example, more than 3500 different Type II restriction enzymes have been characterized.[60] Each enzyme is named after the bacterium from which it was isolated, using a naming system based on bacterial genus, species and strain.[61][62] For example, the name of the EcoRI restriction enzyme was derived as shown in the box.
التطبيقات
Isolated restriction enzymes are used to manipulate DNA for different scientific applications.
They are used to assist insertion of genes into plasmid vectors during gene cloning and protein production experiments. For optimal use, plasmids that are commonly used for gene cloning are modified to include a short polylinker sequence (called the multiple cloning site, or MCS) rich in restriction enzyme recognition sequences. This allows flexibility when inserting gene fragments into the plasmid vector; restriction sites contained naturally within genes influence the choice of endonuclease for digesting the DNA, since it is necessary to avoid restriction of wanted DNA while intentionally cutting the ends of the DNA. To clone a gene fragment into a vector, both plasmid DNA and gene insert are typically cut with the same restriction enzymes, and then glued together with the assistance of an enzyme known as a DNA ligase.[63][64]
Restriction enzymes can also be used to distinguish gene alleles by specifically recognizing single base changes in DNA known as single-nucleotide polymorphisms (SNPs).[65][66] This is however only possible if a SNP alters the restriction site present in the allele. In this method, the restriction enzyme can be used to genotype a DNA sample without the need for expensive gene sequencing. The sample is first digested with the restriction enzyme to generate DNA fragments, and then the different sized fragments separated by gel electrophoresis. In general, alleles with correct restriction sites will generate two visible bands of DNA on the gel, and those with altered restriction sites will not be cut and will generate only a single band. A DNA map by restriction digest can also be generated that can give the relative positions of the genes.[67] The different lengths of DNA generated by restriction digest also produce a specific pattern of bands after gel electrophoresis, and can be used for DNA fingerprinting.
In a similar manner, restriction enzymes are used to digest genomic DNA for gene analysis by Southern blot. This technique allows researchers to identify how many copies (or paralogues) of a gene are present in the genome of one individual, or how many gene mutations (polymorphisms) have occurred within a population. The latter example is called restriction fragment length polymorphism (RFLP).[68]
Artificial restriction enzymes created by linking the FokI DNA cleavage domain with an array of DNA binding proteins or zinc finger arrays, denoted zinc finger nucleases (ZFN), are a powerful tool for host genome editing due to their enhanced sequence specificity. ZFN work in pairs, their dimerization being mediated in-situ through the FokI domain. Each zinc finger array (ZFA) is capable of recognizing 9–12 base pairs, making for 18–24 for the pair. A 5–7 bp spacer between the cleavage sites further enhances the specificity of ZFN, making them a safe and more precise tool that can be applied in humans. A recent Phase I clinical trial of ZFN for the targeted abolition of the CCR5 co-receptor for HIV-1 has been undertaken.[69]
Others have proposed using the bacteria R-M system as a model for devising human anti-viral gene or genomic vaccines and therapies since the RM system serves an innate defense-role in bacteria by restricting tropism by bacteriophages.[70] There is research on REases and ZFN that can cleave the DNA of various human viruses, including HSV-2, high-risk HPVs and HIV-1, with the ultimate goal of inducing target mutagenesis and aberrations of human-infecting viruses.[71][72][73] The human genome already contains remnants of retroviral genomes that have been inactivated and harnessed for self-gain. Indeed, the mechanisms for silencing active L1 genomic retroelements by the three prime repair exonuclease 1 (TREX1) and excision repair cross complementing 1(ERCC) appear to mimic the action of RM-systems in bacteria, and the non-homologous end-joining (NHEJ) that follows the use of ZFN without a repair template.[74][75]
أمثلة
Examples of restriction enzymes include:[76]
| Enzyme | Source | Recognition Sequence | Cut |
|---|---|---|---|
| EcoRI | Escherichia coli |
5'GAATTC 3'CTTAAG |
5'---G AATTC---3' 3'---CTTAA G---5' |
| EcoRII | Escherichia coli |
5'CCWGG 3'GGWCC |
5'--- CCWGG---3' 3'---GGWCC ---5' |
| BamHI | Bacillus amyloliquefaciens |
5'GGATCC 3'CCTAGG |
5'---G GATCC---3' 3'---CCTAG G---5' |
| HindIII | Haemophilus influenzae |
5'AAGCTT 3'TTCGAA |
5'---A AGCTT---3' 3'---TTCGA A---5' |
| TaqI | Thermus aquaticus |
5'TCGA 3'AGCT |
5'---T CGA---3' 3'---AGC T---5' |
| NotI | Nocardia otitidis |
5'GCGGCCGC 3'CGCCGGCG |
5'---GC GGCCGC---3' 3'---CGCCGG CG---5' |
| HinFI | Haemophilus influenzae |
5'GANTC 3'CTNAG |
5'---G ANTC---3' 3'---CTNA G---5' |
| Sau3AI | Staphylococcus aureus |
5'GATC 3'CTAG |
5'--- GATC---3' 3'---CTAG ---5' |
| PvuII* | Proteus vulgaris |
5'CAGCTG 3'GTCGAC |
5'---CAG CTG---3' 3'---GTC GAC---5' |
| SmaI* | Serratia marcescens |
5'CCCGGG 3'GGGCCC |
5'---CCC GGG---3' 3'---GGG CCC---5' |
| HaeIII* | Haemophilus aegyptius |
5'GGCC 3'CCGG |
5'---GG CC---3' 3'---CC GG---5' |
| HgaI[77] | Haemophilus gallinarum |
5'GACGC 3'CTGCG |
5'---NN NN---3' 3'---NN NN---5' |
| AluI* | Arthrobacter luteus |
5'AGCT 3'TCGA |
5'---AG CT---3' 3'---TC GA---5' |
| EcoRV* | Escherichia coli |
5'GATATC 3'CTATAG |
5'---GAT ATC---3' 3'---CTA TAG---5' |
| EcoP15I | Escherichia coli |
5'CAGCAGN25NN 3'GTCGTCN25NN |
5'---CAGCAGN25 NN---3' 3'---GTCGTCN25NN ---5' |
| KpnI[78] | Klebsiella pneumoniae |
5'GGTACC 3'CCATGG |
5'---GGTAC C---3' 3'---C CATGG---5' |
| PstI[78] | Providencia stuartii |
5'CTGCAG 3'GACGTC |
5'---CTGCA G---3' 3'---G ACGTC---5' |
| SacI[78] | Streptomyces achromogenes |
5'GAGCTC 3'CTCGAG |
5'---GAGCT C---3' 3'---C TCGAG---5' |
| SalI[78] | Streptomyces albus |
5'GTCGAC 3'CAGCTG |
5'---G TCGAC---3' 3'---CAGCT G---5' |
| ScaI*[78] | Streptomyces caespitosus |
5'AGTACT 3'TCATGA |
5'---AGT ACT---3' 3'---TCA TGA---5' |
| SpeI | Sphaerotilus natans |
5'ACTAGT 3'TGATCA |
5'---A CTAGT---3' 3'---TGATC A---5' |
| SphI[78] | Streptomyces phaeochromogenes |
5'GCATGC 3'CGTACG |
5'---GCATG C---3' 3'---C GTACG---5' |
| StuI*[79][80] | Streptomyces tubercidicus |
5'AGGCCT 3'TCCGGA |
5'---AGG CCT---3' 3'---TCC GGA---5' |
| XbaI[78] | Xanthomonas badrii |
5'TCTAGA 3'AGATCT |
5'---T CTAGA---3' 3'---AGATC T---5' |
Key:
* = blunt ends
N = C or G or T or A
W = A or T
انظر أيضا
- Detailed articles about certain restriction enzymes: EcoRI, HindIII, BglII.
- List of restriction enzyme cutting sites
- Homing endonuclease
- List of homing endonuclease cutting sites
- Isoschizomer.
- Star activity
- Molecular weight size marker
الهامش
- ^ Roberts RJ (November 1976). "Restriction endonucleases". CRC Critical Reviews in Biochemistry. 4 (2): 123–64. doi:10.3109/10409237609105456. PMID 795607.
- ^ أ ب Kessler C, Manta V (August 1990). "Specificity of restriction endonucleases and DNA modification methyltransferases a review (Edition 3)". Gene. 92 (1–2): 1–248. doi:10.1016/0378-1119(90)90486-B. PMID 2172084.
- ^ Pingoud A, Alves J, Geiger R (1993). "Chapter 8: Restriction Enzymes". In Burrell M (ed.). Enzymes of Molecular Biology. Methods of Molecular Biology. Vol. 16. Totowa, NJ: Humana Press. pp. 107–200. ISBN 0-89603-234-5.
- ^ أ ب Arber W, Linn S (1969). "DNA modification and restriction". Annual Review of Biochemistry. 38: 467–500. doi:10.1146/annurev.bi.38.070169.002343. PMID 4897066.
- ^ Krüger DH, Bickle TA (September 1983). "Bacteriophage survival: multiple mechanisms for avoiding the deoxyribonucleic acid restriction systems of their hosts". Microbiological Reviews. 47 (3): 345–60. doi:10.1128/MMBR.47.3.345-360.1983. PMC 281580. PMID 6314109.
- ^ Kobayashi I (September 2001). "Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution". Nucleic Acids Research. 29 (18): 3742–56. doi:10.1093/nar/29.18.3742. PMC 55917. PMID 11557807.
- ^ Roberts RJ (April 2005). "How restriction enzymes became the workhorses of molecular biology". Proceedings of the National Academy of Sciences of the United States of America. 102 (17): 5905–8. Bibcode:2005PNAS..102.5905R. doi:10.1073/pnas.0500923102. PMC 1087929. PMID 15840723.
- ^ Roberts RJ, Vincze T, Posfai J, Macelis D (January 2007). "REBASE--enzymes and genes for DNA restriction and modification". Nucleic Acids Research. 35 (Database issue): D269-70. doi:10.1093/nar/gkl891. PMC 1899104. PMID 17202163.
- ^ Primrose SB, Old RW (1994). Principles of gene manipulation: an introduction to genetic engineering. Oxford: Blackwell Scientific. ISBN 0-632-03712-1.
- ^ Micklos DA, Bloom MV, Freyer GA (1996). Laboratory DNA science: an introduction to recombinant DNA techniques and methods of genome analysis. Menlo Park, Calif: Benjamin/Cummings Pub. Co. ISBN 0-8053-3040-2.
- ^ Massey A, Kreuzer H (2001). Recombinant DNA and Biotechnology: A Guide for Students. Washington, D.C: ASM Press. ISBN 1-55581-176-0.
- ^ Winnacker E-L (1987). "Chapter 2: Isolation, Identification, and Characterisation of DNA fragments". From Genes to Clones. VCH. ISBN 0-89573-614-4.
- ^ أ ب Luria SE, Human ML (October 1952). "A nonhereditary, host-induced variation of bacterial viruses". Journal of Bacteriology. 64 (4): 557–69. doi:10.1128/JB.64.4.557-569.1952. PMC 169391. PMID 12999684.
- ^ Bertani G, Weigle JJ (February 1953). "Host controlled variation in bacterial viruses". Journal of Bacteriology. 65 (2): 113–21. doi:10.1128/JB.65.2.113-121.1953. PMC 169650. PMID 13034700.
- ^ Meselson M, Yuan R (March 1968). "DNA restriction enzyme from E. coli". Nature. 217 (5134): 1110–4. Bibcode:1968Natur.217.1110M. doi:10.1038/2171110a0. PMID 4868368. S2CID 4172829.
- ^ Dussoix D, Arber W (July 1962). "Host specificity of DNA produced by Escherichia coli. II. Control over acceptance of DNA from infecting phage lambda". Journal of Molecular Biology. 5 (1): 37–49. doi:10.1016/S0022-2836(62)80059-X. PMID 13888713.
- ^ Lederberg S, Meselson M (May 1964). "Degradation of non-replicating bacteriophage dna in non-accepting cells". Journal of Molecular Biology. 8 (5): 623–8. doi:10.1016/S0022-2836(64)80112-1. PMID 14187389.
- ^ Roberts RJ (April 2005). "How restriction enzymes became the workhorses of molecular biology". Proceedings of the National Academy of Sciences of the United States of America. 102 (17): 5905–8. Bibcode:2005PNAS..102.5905R. doi:10.1073/pnas.0500923102. PMC 1087929. PMID 15840723.
- ^ Smith HO, Wilcox KW (July 1970). "A restriction enzyme from Hemophilus influenzae. I. Purification and general properties". Journal of Molecular Biology. 51 (2): 379–91. doi:10.1016/0022-2836(70)90149-X. PMID 5312500.
- ^ Kelly TJ, Smith HO (July 1970). "A restriction enzyme from Hemophilus influenzae. II". Journal of Molecular Biology. 51 (2): 393–409. doi:10.1016/0022-2836(70)90150-6. PMID 5312501.
- ^ Loenen WA, Dryden DT, Raleigh EA, Wilson GG, Murray NE (January 2014). "Highlights of the DNA cutters: a short history of the restriction enzymes". Nucleic Acids Research. 42 (1): 3–19. doi:10.1093/nar/gkt990. PMC 3874209. PMID 24141096.
- ^ Danna K, Nathans D (December 1971). "Specific cleavage of simian virus 40 DNA by restriction endonuclease of Hemophilus influenzae". Proceedings of the National Academy of Sciences of the United States of America. 68 (12): 2913–7. Bibcode:1971PNAS...68.2913D. doi:10.1073/pnas.68.12.2913. PMC 389558. PMID 4332003.
- ^ "The Nobel Prize in Physiology or Medicine". The Nobel Foundation. 1978. Retrieved 2008-06-07.
for the discovery of restriction enzymes and their application to problems of molecular genetics
- ^ Villa-Komaroff L, Efstratiadis A, Broome S, Lomedico P, Tizard R, Naber SP, et al. (August 1978). "A bacterial clone synthesizing proinsulin". Proceedings of the National Academy of Sciences of the United States of America. 75 (8): 3727–31. Bibcode:1978PNAS...75.3727V. doi:10.1073/pnas.75.8.3727. PMC 392859. PMID 358198.
- ^ Jeltsch A, Kröger M, Pingoud A (July 1995). "Evidence for an evolutionary relationship among type-II restriction endonucleases". Gene. 160 (1): 7–16. doi:10.1016/0378-1119(95)00181-5. PMID 7628720.
- ^ Jeltsch A, Pingoud A (February 1996). "Horizontal gene transfer contributes to the wide distribution and evolution of type II restriction-modification systems". Journal of Molecular Evolution. 42 (2): 91–6. Bibcode:1996JMolE..42...91J. doi:10.1007/BF02198833. PMID 8919860. S2CID 19989648.
- ^ Naito T, Kusano K, Kobayashi I (February 1995). "Selfish behavior of restriction-modification systems". Science. 267 (5199): 897–9. Bibcode:1995Sci...267..897N. doi:10.1126/science.7846533. PMID 7846533. S2CID 31128438.
- ^ Cooper S (2003). "Restriction Map". bioweb.uwlax.edu. University of Wisconsin. Retrieved 10 May 2021.
- ^ أ ب ت ث ج ح خ د ذ ر Pingoud A, Jeltsch A (September 2001). "Structure and function of type II restriction endonucleases". Nucleic Acids Research. 29 (18): 3705–27. doi:10.1093/nar/29.18.3705. PMC 55916. PMID 11557805.
- ^ Clark DP (2005). Molecular biology. Amsterdam: Elsevier Academic Press. ISBN 0-12-175551-7.
- ^ Goodsell DS (2002). "The molecular perspective: restriction endonucleases" (PDF). Stem Cells. 20 (2): 190–1. doi:10.1634/stemcells.20-2-190. PMID 11897876. S2CID 222199041.
- ^ أ ب Bickle TA, Krüger DH (June 1993). "Biology of DNA restriction". Microbiological Reviews. 57 (2): 434–50. doi:10.1128/MMBR.57.2.434-450.1993. PMC 372918. PMID 8336674.
- ^ Boyer HW (1971). "DNA restriction and modification mechanisms in bacteria". Annual Review of Microbiology. 25: 153–76. doi:10.1146/annurev.mi.25.100171.001101. PMID 4949033.
- ^ Yuan R (1981). "Structure and mechanism of multifunctional restriction endonucleases". Annual Review of Biochemistry. 50: 285–319. doi:10.1146/annurev.bi.50.070181.001441. PMID 6267988.
- ^ أ ب Types of Restriction Endonucleases | NEB
- ^ أ ب Sistla S, Rao DN (2004). "S-Adenosyl-L-methionine-dependent restriction enzymes". Critical Reviews in Biochemistry and Molecular Biology. 39 (1): 1–19. doi:10.1080/10409230490440532. PMID 15121719. S2CID 1929381.
- ^ Williams RJ (March 2003). "Restriction endonucleases: classification, properties, and applications". Molecular Biotechnology. 23 (3): 225–43. doi:10.1385/MB:23:3:225. PMID 12665693. S2CID 29672999.
- ^ أ ب Murray NE (June 2000). "Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle)". Microbiology and Molecular Biology Reviews. 64 (2): 412–34. doi:10.1128/MMBR.64.2.412-434.2000. PMC 98998. PMID 10839821.
- ^ PDB 1qps Gigorescu A, Morvath M, Wilkosz PA, Chandrasekhar K, Rosenberg JM (2004). "The integration of recognition and cleavage: X-ray structures of pre-transition state complex, post-reactive complex, and the DNA-free endonuclease". In Alfred M. Pingoud (ed.). Restriction Endonucleases (Nucleic Acids and Molecular Biology, Volume 14). Berlin: Springer. pp. 137–178. ISBN 3-540-20502-0.
- ^ Ninfa JP, Balou DP, Benore M (2010). Fundamental Laboratory Approaches for Biochemistry and Biotechnology. Hoboken, NJ: John Wiley & Sons. p. 341. ISBN 978-0-470-08766-4.
- ^ Siwek W, Czapinska H, Bochtler M, Bujnicki JM, Skowronek K (August 2012). "Crystal structure and mechanism of action of the N6-methyladenine-dependent type IIM restriction endonuclease R.DpnI". Nucleic Acids Research. 40 (15): 7563–72. doi:10.1093/nar/gks428. PMC 3424567. PMID 22610857.
- ^ Mierzejewska K, Siwek W, Czapinska H, Kaus-Drobek M, Radlinska M, Skowronek K, et al. (July 2014). "Structural basis of the methylation specificity of R.DpnI". Nucleic Acids Research. 42 (13): 8745–54. doi:10.1093/nar/gku546. PMC 4117772. PMID 24966351.
- ^ Dryden DT, Murray NE, Rao DN (September 2001). "Nucleoside triphosphate-dependent restriction enzymes". Nucleic Acids Research. 29 (18): 3728–41. doi:10.1093/nar/29.18.3728. PMC 55918. PMID 11557806.
- ^ Meisel A, Bickle TA, Krüger DH, Schroeder C (January 1992). "Type III restriction enzymes need two inversely oriented recognition sites for DNA cleavage". Nature. 355 (6359): 467–9. Bibcode:1992Natur.355..467M. doi:10.1038/355467a0. PMID 1734285. S2CID 4354056.
- ^ Bourniquel AA, Bickle TA (November 2002). "Complex restriction enzymes: NTP-driven molecular motors". Biochimie. 84 (11): 1047–59. doi:10.1016/S0300-9084(02)00020-2. PMID 12595133.
- ^ أ ب Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. (March 2007). "CRISPR provides acquired resistance against viruses in prokaryotes". Science. 315 (5819): 1709–12. Bibcode:2007Sci...315.1709B. doi:10.1126/science.1138140. hdl:20.500.11794/38902. PMID 17379808. S2CID 3888761.
- ^ Horvath P, Barrangou R (January 2010). "CRISPR/Cas, the immune system of bacteria and archaea". Science. 327 (5962): 167–70. Bibcode:2010Sci...327..167H. doi:10.1126/science.1179555. PMID 20056882. S2CID 17960960.
- ^ Kim YG, Cha J, Chandrasegaran S (February 1996). "Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain". Proceedings of the National Academy of Sciences of the United States of America. 93 (3): 1156–60. Bibcode:1996PNAS...93.1156K. doi:10.1073/pnas.93.3.1156. PMC 40048. PMID 8577732.
- ^ Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD (September 2010). "Genome editing with engineered zinc finger nucleases". Nature Reviews. Genetics. 11 (9): 636–46. doi:10.1038/nrg2842. PMID 20717154. S2CID 205484701.
- ^ Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, Voytas DF (May 2009). "High-frequency modification of plant genes using engineered zinc-finger nucleases". Nature. 459 (7245): 442–5. Bibcode:2009Natur.459..442T. doi:10.1038/nature07845. PMC 2743854. PMID 19404258.
- ^ Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, et al. (May 2009). "Precise genome modification in the crop species Zea mays using zinc-finger nucleases". Nature. 459 (7245): 437–41. Bibcode:2009Natur.459..437S. doi:10.1038/nature07992. PMID 19404259. S2CID 4323298.
- ^ Ekker SC (2008). "Zinc finger-based knockout punches for zebrafish genes". Zebrafish. 5 (2): 121–3. doi:10.1089/zeb.2008.9988. PMC 2849655. PMID 18554175.
- ^ Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, et al. (July 2009). "Knockout rats via embryo microinjection of zinc-finger nucleases". Science. 325 (5939): 433. Bibcode:2009Sci...325..433G. doi:10.1126/science.1172447. PMC 2831805. PMID 19628861.
- ^ Tovkach A, Zeevi V, Tzfira T (January 2011). "Expression, purification and characterization of cloning-grade zinc finger nuclease". Journal of Biotechnology. 151 (1): 1–8. doi:10.1016/j.jbiotec.2010.10.071. PMID 21029755.
- ^ Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, et al. (October 2010). "Targeting DNA double-strand breaks with TAL effector nucleases". Genetics. 186 (2): 757–61. doi:10.1534/genetics.110.120717. PMC 2942870. PMID 20660643.
- ^ Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, Yang B (January 2011). "TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain". Nucleic Acids Research. 39 (1): 359–72. doi:10.1093/nar/gkq704. PMC 3017587. PMID 20699274.
- ^ Hsu PD, Lander ES, Zhang F (June 2014). "Development and applications of CRISPR-Cas9 for genome engineering". Cell. 157 (6): 1262–78. doi:10.1016/j.cell.2014.05.010. PMC 4343198. PMID 24906146.
- ^ "Revolutionizing Biotechnology with Artificial Restriction Enzymes". Genetic Engineering and Biotechnology News. 10 February 2017. Retrieved 27 May 2021. (reporting on Programmable DNA-Guided Artificial Restriction Enzymes)
- ^ Murtola M, Wenska M, Strömberg R (July 2010). "PNAzymes that are artificial RNA restriction enzymes". Journal of the American Chemical Society. 132 (26): 8984–90. doi:10.1021/ja1008739. PMID 20545354.
- ^ A. Pingoud (2004). Restriction Endonucleases (Nucleic Acids and Molecular Biology). Springer. p. 3. ISBN 9783642188510.
- ^ Smith HO, Nathans D (December 1973). "Letter: A suggested nomenclature for bacterial host modification and restriction systems and their enzymes". Journal of Molecular Biology. 81 (3): 419–23. doi:10.1016/0022-2836(73)90152-6. PMID 4588280.
- ^ Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, et al. (April 2003). "A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes". Nucleic Acids Research. 31 (7): 1805–12. doi:10.1093/nar/gkg274. PMC 152790. PMID 12654995.
- ^ Geerlof A. "Cloning using restriction enzymes". European Molecular Biology Laboratory - Hamburg. Retrieved 2008-06-07.
- ^ Russell DW, Sambrook J (2001). Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory. ISBN 0-87969-576-5.
- ^ Wolff JN, Gemmell NJ (February 2008). "Combining allele-specific fluorescent probes and restriction assay in real-time PCR to achieve SNP scoring beyond allele ratios of 1:1000". BioTechniques. 44 (2): 193–4, 196, 199. doi:10.2144/000112719. PMID 18330346.
- ^ Zhang R, Zhu Z, Zhu H, Nguyen T, Yao F, Xia K, et al. (July 2005). "SNP Cutter: a comprehensive tool for SNP PCR-RFLP assay design". Nucleic Acids Research. 33 (Web Server issue): W489-92. doi:10.1093/nar/gki358. PMC 1160119. PMID 15980518.
- ^ "Mapping". Nature.
- ^ Stryer L, Berg JM, Tymoczko JL (2002). Biochemistry (Fifth ed.). San Francisco: W.H. Freeman. p. 122. ISBN 0-7167-4684-0.
- ^ Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, et al. (March 2014). "Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV". The New England Journal of Medicine. 370 (10): 901–10. doi:10.1056/NEJMoa1300662. PMC 4084652. PMID 24597865.
- ^ Wayengera M (2003). "HIV and Gene Therapy: The proposed [R-M enzymatic] model for a gene therapy against HIV". Makerere Med J. 38: 28–30.
- ^ Wayengera M, Kajumbula H, Byarugaba W (2007). "Frequency and site mapping of HIV-1/SIVcpz, HIV-2/SIVsmm and Other SIV gene sequence cleavage by various bacteria restriction enzymes: Precursors for a novel HIV inhibitory product". Afr J Biotechnol. 6 (10): 1225–1232.
- ^ Schiffer JT, Aubert M, Weber ND, Mintzer E, Stone D, Jerome KR (September 2012). "Targeted DNA mutagenesis for the cure of chronic viral infections". Journal of Virology. 86 (17): 8920–36. doi:10.1128/JVI.00052-12. PMC 3416169. PMID 22718830.
- ^ Manjunath N, Yi G, Dang Y, Shankar P (November 2013). "Newer gene editing technologies toward HIV gene therapy". Viruses. 5 (11): 2748–66. doi:10.3390/v5112748. PMC 3856413. PMID 24284874.
- ^ Stetson DB, Ko JS, Heidmann T, Medzhitov R (August 2008). "Trex1 prevents cell-intrinsic initiation of autoimmunity". Cell. 134 (4): 587–98. doi:10.1016/j.cell.2008.06.032. PMC 2626626. PMID 18724932.
- ^ Gasior SL, Roy-Engel AM, Deininger PL (June 2008). "ERCC1/XPF limits L1 retrotransposition". DNA Repair. 7 (6): 983–9. doi:10.1016/j.dnarep.2008.02.006. PMC 2483505. PMID 18396111.
- ^ Roberts RJ (January 1980). "Restriction and modification enzymes and their recognition sequences". Nucleic Acids Research. 8 (1): r63–r80. doi:10.1093/nar/8.1.197-d. PMC 327257. PMID 6243774.
- ^ Roberts RJ (1988). "Restriction enzymes and their isoschizomers". Nucleic Acids Research. 16 Suppl (Suppl): r271-313. doi:10.1093/nar/16.suppl.r271. PMC 340913. PMID 2835753.
- ^ أ ب ت ث ج ح خ Krieger M, Scott MP, Matsudaira PT, Lodish HF, Darnell JE, Zipursky L, Kaiser C, Berk A (2004). Molecular Cell Biology (5th ed.). New York: W.H. Freeman and Company. ISBN 0-7167-4366-3.
- ^ "Stu I from Streptomyces tubercidicus". Sigma-Aldrich. Retrieved 2008-06-07.
- ^ Shimotsu H, Takahashi H, Saito H (November 1980). "A new site-specific endonuclease StuI from Streptomyces tubercidicus". Gene. 11 (3–4): 219–25. doi:10.1016/0378-1119(80)90062-1. PMID 6260571.
وصلات خارجية
عامة:
- MeSH DNA+Restriction+Enzymes
- Firman K (2007-11-24). "Type I Restriction-Modification". University of Portsmouth. Retrieved 2008-06-06.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help) - Goodsell DS (2000-08-01). "Restriction Enzymes". Molecule of the Month. RCSB Protein Data Bank. Retrieved 2008-06-06.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help) - Simmer M, Secko D (2003-08-01). "Restriction Endonucleases: Molecular Scissors for Specifically Cutting DNA". The Science Creative Quarterly. Retrieved 2008-06-06.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help)
قواعد بيانات:
- Roberts RJ, Vincze T, Posfai, J, Macelis D. "[[Rebase (database)|REBASE]]". Retrieved 2008-06-06.
Restriction Enzyme Database
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help); URL–wikilink conflict (help)CS1 maint: multiple names: authors list (link)
برامج حاسوبية:
- Bikandi J, San Millán R, Rementeria A, and Garaizar J. "Restriction enzyme digest of DNA". insilico.ehu.es. Retrieved 2008-06-06.
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help)CS1 maint: multiple names: authors list (link) - Palmer M. "WatCut". University of Waterloo, Ontario, Canada. Retrieved 2008-06-06.
An on-line tool for restriction analysis, silent mutation scanning, SNP-RFLP analysis
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help) - Vincze,T, Posfai J, Roberts RJ. "NEBcutter V2.0". New England Biolabs Inc. Retrieved 2008-06-06.
Restriction enzyme finder
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help)CS1 maint: multiple names: authors list (link) - "Restriction enzyme digest of DNA software". BioPHP: PHP for Bioinformatics. Retrieved 2008-06-06.
Online tool, free source code
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help) - "pDRAW32". AcaClone software. Retrieved 2008-06-06.
Freeware DNA cloning, sequence analysis and plasmid/DNA plotting software
{{cite web}}: Cite has empty unknown parameter:|coauthors=(help)

